DNA ligase
| DNA ligase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 DNA ligase repairing chromosomal damage | |||||||||
| Identifiers | |||||||||
| EC number | 6.5.1.1 | ||||||||
| CAS number | 9015-85-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
| ligase I, DNA, ATP-dependent | |
|---|---|
 | |
| Identifiers | |
| Symbol | LIG1 |
| Entrez | 3978 |
| HUGO | 6598 |
| OMIM | 126391 |
| RefSeq | NM_000234 |
| UniProt | P18858 |
| Other data | |
| Locus | Chr. 19 |
| ligase III, DNA, ATP-dependent | |
|---|---|
| Identifiers | |
| Symbol | LIG3 |
| Entrez | 3980 |
| HUGO | 6600 |
| OMIM | 600940 |
| RefSeq | NM_002311 |
| UniProt | P49916 |
| Other data | |
| Locus | Chr. 17 q11.2-q12 |
| ligase IV, DNA, ATP-dependent | |
|---|---|
| Identifiers | |
| Symbol | LIG4 |
| Entrez | 3981 |
| HUGO | 6601 |
| OMIM | 601837 |
| RefSeq | NM_002312 |
| UniProt | P49917 |
| Other data | |
| Locus | Chr. 13 q33-q34 |
In molecular biology, DNA ligase is a specific type of enzyme, a ligase, (EC 6.5.1.1) that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living organisms, but some forms (such as DNA ligase IV) may specifically repair double-strand breaks (i.e. a break in both complementary strands of DNA). Single-strand breaks are repaired by DNA ligase using the complementary strand of the double helix as a template,[1] with DNA ligase creating the final phosphodiester bond to fully repair the DNA.
DNA ligase is used in both DNA repair and DNA replication (see Mammalian ligases). In addition, DNA ligase has extensive use in molecular biology laboratories for recombinant DNA experiments (see Applications in molecular biology research). Purified DNA ligase is used in gene cloning to join DNA molecules together to form recombinant DNA.
Ligase mechanism
The mechanism of DNA ligase is to form two covalent phosphodiester bonds between 3' hydroxyl ends of one nucleotide, ("acceptor") with the 5' phosphate end of another ("donor"). ATP is required for the ligase reaction, which proceeds in three steps:
- Adenylation (addition of AMP) of a lysine residue in the active center of the enzyme, pyrophosphate is released;
- Transfer of the AMP to the 5' phosphate of the so-called donor, formation of a pyrophosphate bond;
- Formation of a phosphodiester bond between the 5' phosphate of the donor and the 3' hydroxyl of the acceptor.[2]
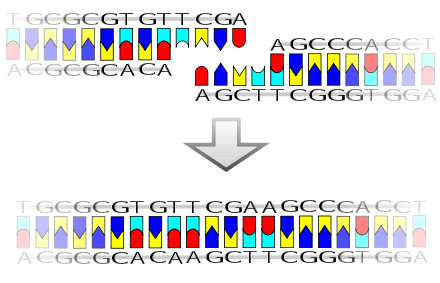
Ligase will also work with blunt ends, although higher enzyme concentrations and different reaction conditions are required.
Types of ligases
E. coli DNA ligase
The E. coli DNA ligase is encoded by the lig gene. DNA ligase in E. coli, as well as most prokaryotes, uses energy gained by cleaving nicotinamide adenine dinucleotide (NAD) to create the phosphodiester bond.[3] It does not ligate blunt-ended DNA except under conditions of molecular crowding with polyethylene glycol, and cannot join RNA to DNA efficiently.
T4 DNA ligase
The DNA ligase from bacteriophage T4 is the ligase most-commonly used in laboratory research.[4] It can ligate cohesive or "sticky" ends of DNA, oligonucleotides, as well as RNA and RNA-DNA hybrids, but not single-stranded nucleic acids. It can also ligate blunt-ended DNA with much greater efficiency than E. coli DNA ligase. Unlike E. coli DNA ligase, T4 DNA ligase cannot utilize NAD and it has an absolute requirement for ATP as a cofactor. Some engineering has been done to improve the in vitro activity of T4 DNA ligase; one successful approach, for example, tested T4 DNA ligase fused to several alternative DNA binding proteins and found that the constructs with either p50 or NF-kB as fusion partners were over 160% more active in blunt-end ligations for cloning purposes than wild type T4 DNA ligase.[5]
Mammalian ligases
In mammals, there are four specific types of ligase.
- DNA ligase I: ligates the nascent DNA of the lagging strand after the Ribonuclease H has removed the RNA primer from the Okazaki fragments.
- DNA ligase III: complexes with DNA repair protein XRCC1 to aid in sealing DNA during the process of nucleotide excision repair and recombinant fragments. Of the all known mammalian DNA ligases, only Lig III has been found to be present in mitochondria.
- DNA ligase IV: complexes with XRCC4. It catalyzes the final step in the non-homologous end joining DNA double-strand break repair pathway. It is also required for V(D)J recombination, the process that generates diversity in immunoglobulin and T-cell receptor loci during immune system development.
- DNA ligase II: appears to be used in repair. It is formed by alternative splicing of a proteolytic fragment of DNA ligase III and does not have its own gene, therefore it is often considered to be virtually identical to DNA ligase III.
DNA ligase from eukaryotes and some microbes uses adenosine triphosphate (ATP) rather than NAD.[3]
Thermostable ligases
Ligases from various thermophilic bacteria have been cloned and sequenced, and are available commercially for use in ligase amplification reaction because of their thermostable properties.
Measurement of ligase activity
There are at least three different units used to measure the activity of DNA ligase:[6]
- Weiss unit - the amount of ligase that catalyzes the exchange of 1 nmole of 32P from inorganic pyrophosphate to ATP in 20 minutes at 37°C. This is the one most commonly used.
- Modrich-Lehman unit - this is rarely used, and one unit is defined as the amount of enzyme required to convert 100 nmoles of d(A-T)n to an exonuclease-III resistant form in 30 minutes under standard conditions.
- Many commercial suppliers of ligases use an arbitrary unit based on the ability of ligase to ligate cohesive ends. These units are often more subjective than quantitative and lack precision.
Applications in molecular biology research
DNA ligases have become indispensable tools in modern molecular biology research for generating recombinant DNA sequences. For example, DNA ligases are used with restriction enzymes to insert DNA fragments, often genes, into plasmids.
Controlling the optimal temperature is a vital aspect of performing efficient recombination experiments involving the ligation of cohesive-ended fragments. Most experiments use T4 DNA Ligase (isolated from bacteriophage T4), which is most active at 37 °C.[7] However, for optimal ligation efficiency with cohesive-ended fragments ("sticky ends"), the optimal enzyme temperature needs to be balanced with the melting temperature Tm of the sticky ends being ligated,[8] the homologous pairing of the sticky ends will not be stable because the high temperature disrupts hydrogen bonding. A ligation reaction is most efficient when the sticky ends are already stably annealed, and disruption of the annealing ends would therefore result in low ligation efficiency. The shorter the overhang, the lower the Tm.
Since blunt-ended DNA fragments have no cohesive ends to anneal, the melting temperature is not a factor to consider within the normal temperature range of the ligation reaction. The limiting factor in blunt end ligation is not the activity of the ligase but rather the number of alignments between DNA fragment ends that occur. The most efficient ligation temperature for blunt-ended DNA would therefore be the temperature at which the greatest number of alignments can occur. The majority of blunt-ended ligations are carried out at 14-25 °C overnight. The absence of stably annealed ends also means that the ligation efficiency is lowered, requiring a higher ligase concentration to be used.[8]
History
The first DNA ligase was purified and characterized in 1967. The common commercially available DNA ligases were originally discovered in bacteriophage T4, E. coli and other bacteria.[9]
See also
References
- ↑ Pascal JM, O'Brien PJ, Tomkinson AE, Ellenberger T (November 2004). "Human DNA ligase I completely encircles and partially unwinds nicked DNA". Nature. 432 (7016): 473–8. doi:10.1038/nature03082. PMID 15565146.
- ↑ Lehman IR (November 1974). "DNA ligase: structure, mechanism, and function". Science. 186 (4166): 790–7. doi:10.1126/science.186.4166.790. PMID 4377758.
- 1 2 Foster JB, Slonczewski J (2010). Microbiology: An Evolving Science (Second ed.). New York: W. W. Norton & Company. ISBN 0-393-93447-0.
- ↑ "Ligases" (PDF). Enzyme Resources Guide. Promega Corporation. pp. 8–14.
- ↑ Wilson RH, Morton SK, Deiderick H, Gerth ML, Paul HA, Gerber I, Patel A, Ellington AD, Hunicke-Smith SP, Patrick WM (July 2013). "Engineered DNA ligases with improved activities in vitro". Protein Eng. Des. Sel. 26 (7): 471–8. doi:10.1093/protein/gzt024. PMID 23754529.
- ↑ Russell DW, Sambrook J (2001). "Chapter 1: Plasmids and Their Usefulness in Molecular Cloning". Molecular cloning: a laboratory manual. 1 (3rd ed.). Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory. pp. 1–159. ISBN 0-87969-577-3.
- ↑ F. Baneyx F; Lucotte G (1993). Introduction to Molecular Cloning Techniques. Chichester: John Wiley & Sons. p. 156. ISBN 978-0471188490.
- 1 2 Tabor S (May 2001). "DNA ligases". Curr Protoc Mol Biol. Chapter 3: Unit 3.14. doi:10.1002/0471142727.mb0314s08. ISBN 0471142727. PMID 18265223.
- ↑ Weiss B, Richardson CC (April 1967). "Enzymatic breakage and joining of deoxyribonucleic acid, I. Repair of single-strand breaks in DNA by an enzyme system from Escherichia coli infected with T4 bacteriophage". Proc. Natl. Acad. Sci. U.S.A. 57 (4): 1021–8. doi:10.1073/pnas.57.4.1021. PMC 224649
 . PMID 5340583.
. PMID 5340583.
External links
- DNA Ligase: PDB molecule of the month
- Davidson College General Information on Ligase
- OpenWetWare DNA Ligation Protocol