bZIP Maf
| bZIP_Maf | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| Symbol | bZIP_Maf | ||||||||
| Pfam | PF03131 | ||||||||
| Pfam clan | CL0018 | ||||||||
| InterPro | IPR004826 | ||||||||
| SCOP | 1k1v | ||||||||
| SUPERFAMILY | 1k1v | ||||||||
| |||||||||
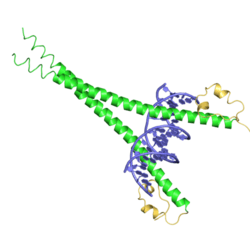
bZIP Maf is a domain found in Maf transcription factor proteins. It contains a leucine zipper (bZIP) domain, which mediates the transcription factor's dimerization and DNA binding properties. The Maf extended homology region (EHR) is present at the N-terminus of the protein. This region (shown in yellow in the adjacent image) exists only within the Maf family and allows the family to recognize longer DNA motifs than other leucine zippers. These motifs are termed the Maf recognition element (MARE) and is 13 or 14 base pairs long. In particular, the two residues at the beginning of helix H2 are positioned to recognise the flanking region of the DNA.[2] Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF2-E2 transcription factor.
In mouse, Maf1 may play an early role in axial patterning. Defects in these proteins are a cause of autosomal dominant retinitis pigmentosa. Neural retina-specific leucine zipper proteins belong to this family.
References
- ↑ Kurokawa, H.; Motohashi, H.; Sueno, S.; Kimura, M.; Takagawa, H.; Kanno, Y.; Yamamoto, M.; Tanaka, T. (2009). "Structural Basis of Alternative DNA Recognition by Maf Transcription Factors". Molecular and Cellular Biology. 29 (23): 6232–6244. doi:10.1128/MCB.00708-09. PMC 2786689
 . PMID 19797082.
. PMID 19797082. - ↑ Kusunoki H, Motohashi H, Katsuoka F, Morohashi A, Yamamoto M, Tanaka T (April 2002). "Solution structure of the DNA-binding domain of MafG". Nat. Struct. Biol. 9 (4): 252–6. doi:10.1038/nsb771. PMID 11875518.
This article incorporates text from the public domain Pfam and InterPro IPR004826