Limb development
| Development of the limbs | |
|---|---|
 Illustration of a human embryo at six weeks gestational age | |
 9-Week Human Embryo from Ectopic Pregnancy | |
Limb development in tetrapods — animals with four limbs — is an area of active research in developmental biology. Limb formation begins in the limb field, as mesenchymal cells from the lateral plate mesoderm proliferate to the point that they cause the ectoderm above to bulge out, which is known as the limb bud. Fibroblast growth factor (FGF) induces formation of an organizer, called the apical ectodermal ridge (AER), which guides further development and controls cell death. Apoptosis — programmed cell death — is necessary to eliminate webbing between digits.
The limb field is a region specified by expression of certain homeobox (Hox) genes and Tbx genes; Tbx5 for forelimb and Tbx4 for hindlimb. Establishment of the limb field requires retinoic acid signaling in the developing trunk of the embryo from which the limb emerges.[1][2]
Limb formation results from series of epithelial-mesenchymal inductions between the mesenchymal cells of the lateral plate mesoderm and the overlying ectodermal cells. Cells from the lateral plate mesoderm and the myotome migrate to the limb field and proliferate to create the limb bud. The lateral plate cells produce the cartilaginous and skeletal portions of the limb while the myotome cells produce the muscle components. The lateral plate mesodermal cells secrete a fibroblast growth factor (FGF7 and FGF10, presumably) to induce the overlying ectoderm to form an important organizing structure called the apical ectodermal ridge (AER).[3] The AER reciprocatively secretes FGF8 and FGF4 which maintains the FGF10 signal and induces proliferation in the mesoderm. The position of FGF10 expression is regulated by Wnt8c in the hindlimb and Wnt2b in the forelimb. The forelimb and the hindlimb are specified by their position along the anterior/posterior axis and possibly by two T-box containing transcription factors: Tbx5 and Tbx4, respectively.[4][5]
Programmed cell death removes the "webbing" between the digits and joints. BMP signaling induces cell death and Noggin blocks cell death in the digits.
Precartilage condensations
The limb's skeletal elements are prefigured by tight aggregates of mesenchymal cells called precartilage condensations. Mesenchymal condensation is mediated by extracellular matrix and cell adhesion molecules.[6] Cartilage differentiates from the condensations, giving rise to the skeletal primordia. In most tetrapod limb skeletons (though not in some amphibians), the cartilage skeleton is replaced by bone later in development.
Periodicities of the limb pattern
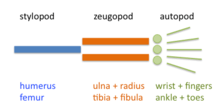
The limb is organized into three regions: stylopod, zeugopod, and autopod (in order from proximal to distal). The two distal regions of the skeleton contain a number of periodic and quasi-periodic pattern motifs. The zeugopod consists of two parallel elements along the anteroposterior (AP) axis and the autopod contains 3-5 (in most cases) elements along the same axis. The digits also have a quasi-periodic arrangement along the proximodistal (PD) axis, consisting of tandem chains of skeletal elements. The generation of the basic limb plan during development results from the patterning of the mesenchyme by an interplay of factors that promote precartilage condensation and factors that inhibit it.[7]
The development of the basic limb plan is accompanied by generation of local differences between the elements. For example, the radius and ulna of the forelimb zeugopod are distinct from one another, as are the different fingers in the autopod. These differences can be treated schematically by considering how they are reflected in each of the limb's three main axes.
Axial patterning and related issues
Many investigations into the development of the limb skeletal pattern have been influenced by the "positional information" concept proposed by Lewis Wolpert in 1971.[8] In tune with this idea, efforts have been made to identify diffusive signaling molecules (morphogens) that traverse orthogonal axes of developing limbs and determine locations and identities of skeletal elements in a concentration-dependent fashion.
Proximal/distal patterning
Hox genes contribute to the specification of the stylopod, zeugopod and autopod. Mutations in Hox genes lead to proximal/distal losses or abnormalities.[9] Three different models have been advanced for explaining the patterning of these regions.
Progress Zone model
The AER creates a zone of cell proliferation and lays down the limb from proximal to distal. The time cells leave the AER determines their positional value. Proximal structures are formed earlier than distal structures.
The Progress Zone model was proposed 30 years ago but recent evidence has conflicted with this model.[10]
Experimental evidence:
- Removing the AER at a later period of development results in less disruption of distal structures than if the AER was removed early in development.
- Grafting an early limb bud tip onto a late wing results in duplication of structures, while grafting a late wing bud tip onto an early limb results in a deletion of structures.
Early allocation and progenitor expansion model (or prespecification model)
Cells are specified for each segment in the early limb bud and this population of cells expand out as the limb bud grows. This model is consistent with the following observations. Cell division is seen throughout the limb bud. Cell death occurs within a 200 μm zone subjacent to the AER when it is removed; cell death forecloses some patterning. FGF-releasing beads are able to rescue limb development when the AER is removed by preventing this cell death.
Experimental evidence:
- Labeled cells in different position of an early limb bud were restricted to single segments of the limb.[11]
- Limbs lacking expression of required FGF4 & FGF8 showed all structures of the limb and not just the proximal parts.[12]
More recently, however, the investigators primarily responsible for both the Progress Zone and Prespecification models have acknowledged that neither of these models accounts adequately for the available experimental data.[10]
Reaction-diffusion (or Turing-type) model
This model, first proposed in 1979,[13] is based on the self-organizing properties of excitable media described by Alan Turing in 1952.[14] The excitable medium is the limb bud mesenchyme, in which cells interact by positively autoregulatory morphogens such as TGF-β and inhibitory signaling pathways involving FGF and Notch. Proximodistal and anteroposterior axes are not considered to be independently specified, but instead emerge by transitions in the number of parallel elements as the undifferentiated apical zone of the growing limb bud undergoes reshaping.[15] This model only specifies a "bare bones" pattern. Other factors like Sonic hedgehog (Shh) and Hox proteins, primary informational molecules in the other models, are proposed instead to play a fine-tuning role.
Experimental evidence:
- Limb mesenchymal cells, when dissociated and grown in culture or reintroduced within ectodermal "hulls" can recapitulate essential aspects of pattern formation, morphogenesis and differentiation.[16][17]
- Peculiarities of the limb skeletal pattern in the mouse Doublefoot mutant are predicted outcomes of a Turing-type mechanism.[18]
- Progressive reduction in distal Hox genes in a Gli3-null background results in progressively more severe polydactyly, displaying thinner and densely packed digits, suggesting (with the aid of computer modeling) that the dose of distal Hox genes modulates the period or wavelength of digits specified by a Turing-type mechanism.[19]
Anterior/posterior patterning
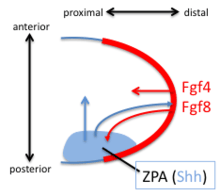
The Zone of Polarizing Activity (ZPA) in the limb bud has pattern-organizing activity by action of a morphogen gradient of Sonic hedgehog (Shh). Shh is both sufficient and necessary to create the ZPA and specify the anterior/posterior pattern in the distal limb (Shh is not necessary for the polarity of the stylopod). Shh is turned on in the posterior through the early expression of Hoxd genes, the expression of Hoxb8, and the expression dHAND. Shh is maintained in the posterior though a feedback loop between the ZPA and the AER. Shh induces the AER to produce FGF4 and FGF8 which maintains the expression of Shh.
Digits 3,4 and 5 are specified by a temporal gradient of Shh. Digit 2 is specified by a long-range diffusible form of Shh and Digit 1 does not require Shh. Shh cleaves the Ci/Gli3 transcriptional repressor complex to convert the transcription factor Gli3 to an activator which activates the transcription of HoxD genes along the anterior/posterior axis. Loss of the Gli3 repressor leads to the formation of generic (non-individualized) digits in extra quantities.[20]
Dorsal/ventral patterning
Dorsal/Ventral patterning arises from Wnt7a signals in the overlying ectoderm not the mesoderm. Wnt7a is both necessary and sufficient to dorsalize the limb. Wnt7a also influences the anterior/posterior axis and loss of Wnt7a causes the dorsal side of limbs to become ventral sides and causes missing posterior digits. Replacing Wnt7a signals rescues this defect. Wnt7a is also required to maintain expression of Shh.
Wnt-7a also causes Lmx-1, a Lim Hox gene (and thus a transcription factor), to be expressed. Lmx-1 is involved in dorsalisation of the limb, which was shown by knocking out the Lmx-1 gene in mice.[21] The mice lacking the Lmx-1 produced ventral skin on both sides of their paws. There are other factors thought to control the DV patterning; Engrailed-1 represses the dorsalizing effect of Wnt-7a on the ventral side of the limbs.[22]
References
- ↑ Stratford T, Horton C, Maden M (1996). "Retinoic acid is required for the initiation of outgrowth in the chick limb bud". Curr Biol. 6 (9): 1124–33. doi:10.1016/S0960-9822(02)70679-9. PMID 8805369.
- ↑ Zhao X, Sirbu IO, Mic FA, et al. (June 2009). "Retinoic acid promotes limb induction through effects on body axis extension but is unnecessary for limb patterning". Curr. Biol. 19 (12): 1050–7. doi:10.1016/j.cub.2009.04.059. PMC 2701469
 . PMID 19464179.
. PMID 19464179. - ↑ Yonei-Tamura S, Endo T, Yajima H, Ohuchi H, Ide H, Tamura K (1999). "FGF7 and FGF10 directly induce the apical ectodermal ridge in chick embryos". Dev Biol. 211 (1): 133–43. doi:10.1006/dbio.1999.9290. PMID 10373311.
- ↑ Ohuchi H, Takeuchi J, Yoshioka H, Ishimaru Y, Ogura K, Takahashi N, Ogura T, Noji S (1998). "Correlation of wing-leg identity in ectopic FGF-induced chimeric limbs with the differential expression of chick Tbx5 and Tbx4". Development. 125 (1): 51–60. PMID 9389663.
- ↑ Rodriguez-Esteban C, Tsukui T, Yonei S, Magallon J, Tamura K, Izpisua Belmonte JC (1999). "The T-box genes Tbx4 and Tbx5 regulate limb outgrowth and identity". Nature. 398 (6730): 814–8. doi:10.1038/19769. PMID 10235264.
- ↑ Hall BK, Miyake T (2000). "All for one and one for all: condensations and the initiation of skeletal development". BioEssays. 22 (2): 138–47. doi:10.1002/(SICI)1521-1878(200002)22:2<138::AID-BIES5>3.0.CO;2-4. PMID 10655033.
- ↑ Newman SA, Bhat R (2007). "Activator-inhibitor dynamics of vertebrate limb pattern formation". Birth Defects Res C Embryo Today. 81 (4): 305–19. doi:10.1002/bdrc.20112. PMID 18228262.
- ↑ Wolpert L (1971). "Positional information and pattern formation". Curr Top Dev Biol. 6 (6): 183–224. doi:10.1016/S0070-2153(08)60641-9. PMID 4950136.
- ↑ Wellik D, Capecchi M (2003). "Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton". Science. 301 (5631): 363–7. doi:10.1126/science.1085672. PMID 12869760.
- 1 2 Tabin C, Wolpert L (2007). "Rethinking the proximodistal axis of the vertebrate limb in the molecular era". Genes Dev. 21 (12): 1433–42. doi:10.1101/gad.1547407. PMID 17575045.
- ↑ Dudley A, Ros M, Tabin C (2002). "A re-examination of proximodistal patterning during vertebrate limb development". Nature. 418 (6897): 539–44. doi:10.1038/nature00945. PMID 12152081.
- ↑ Sun X, Mariani F, Martin G (2002). "Functions of FGF signalling from the apical ectodermal ridge in limb development". Nature. 418 (6897): 501–8. doi:10.1038/nature00902. PMID 12152071.
- ↑ Newman SA, Frisch HL (1979). "Dynamics of skeletal pattern formation in developing chick limb". Science. 205 (4407): 662–668. doi:10.1126/science.462174. PMID 462174.
- ↑ Turing, AM (1952). "The chemical basis of morphogenesis". Philosophical Transactions of the Royal Society B. 237 (641): 37–72. doi:10.1098/rstb.1952.0012.
- ↑ Zhu J, Zhang YT, Alber MS, Newman SA (2010). "Bare bones pattern formation: a core regulatory network in varying geometries reproduces major features of vertebrate limb development and evolution". PLoS ONE. 5 (5): e:10892. doi:10.1371/journal.pone.0010892. PMC 2878345
 . PMID 20531940.
. PMID 20531940. - ↑ Moftah MZ, Downie SA, Bronstein NB, Mezentseva N, Pu J, Maher PA, Newman SA (2002). "Ectodermal FGFs induce perinodular inhibition of limb chondrogenesis in vitro and in vivo via FGF receptor 2". Dev Biol. 249 (2): 270–82. doi:10.1006/dbio.2002.0766. PMID 12221006.
- ↑ Ros MA, Lyons GE, Mackem S, Fallon JF (1994). "Recombinant limbs as a model to study homeobox gene regulation during limb development". Dev Biol. 166 (1): 59–72. doi:10.1006/dbio.1994.1296. PMID 7958460.
- ↑ Miura T, Shiota K, Morriss-Kay G, Maini PK (2006). "Mixed-mode pattern in Doublefoot mutant mouse limb−Turing reaction-diffusion model on a growing domain during limb development". J Theor Biol. 240 (4): 562–73. doi:10.1016/j.jtbi.2005.10.016. PMID 16364368.
- ↑ Sheth R, Marcon L, Bastida MF, Junco M, Quintana L, Dahn R, Kmita M, Sharpe J, Ros MA (2012). "Hox genes regulate digit patterning by controlling the wavelength of a Turing-type mechanism". Science. 338 (6113): 1476–1480. doi:10.1126/science.1226804. PMID 23239739.
- ↑ Chiang C, Litingtung Y, Harris M, Simandl B, Li Y, Beachy P, Fallon J (2001). "Manifestation of the limb prepattern: limb development in the absence of sonic hedgehog function". Dev Biol. 236 (2): 421–35. doi:10.1006/dbio.2001.0346. PMID 11476582.
- ↑ Riddle RD, Ensini M, Nelson C, Tsuchida T, Jessell TM, Tabin C (1995). "'Induction of the LIM homeobox gene Lmx1 by WNT7a establishes dorsoventral pattern in the vertebrate limb". Cell. 17;83(4) (2): 631–40. doi:10.1016/0092-8674(95)90103-5. PMID 7585966.
- ↑ http://genetics.med.harvard.edu/~tabin/Pdfs/Johnson.pdf